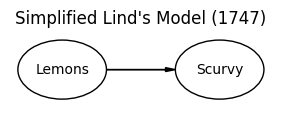
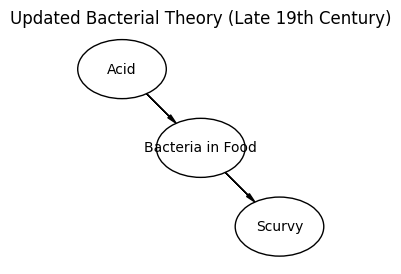
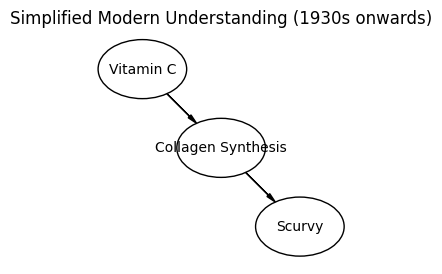
This challenge pushes boundaries intentionally. You’ll tackle problems that normally require weeks of study, but with Cursor AI as your partner (and your brain keeping it honest), you can accomplish more than you thought possible.
The new reality: The four stages of competence are Ignorance → Awareness → Learning → Mastery. AI lets us produce Mastery-level work while operating primarily in the Awareness stage. I focus on awareness training, you leverage AI for execution, and together we create outputs that used to require years of dedicated study.